New Biosensor with Dual-Mode Fluorescence and Electrochemistry
Recently, researchers from the Suzhou Institute of Biomedical Engineering and Technology (SIBET) of the Chinese Academy of Sciences proposed a technique for hand-in-hand structured DNA construction.
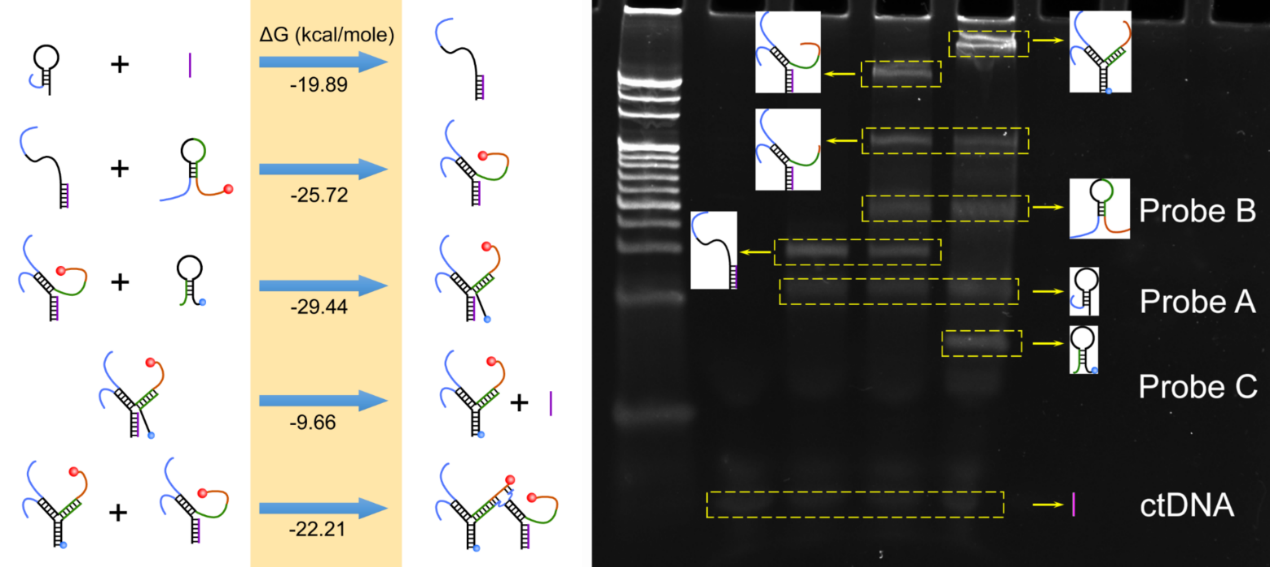
PAGE analysis and reaction process standard free energy (G) Suzhou Institute of Biomedical Engineering and Technology, source of the image (SIBET)
Additionally, they have developed an electrochemical/fluorescent dual-mode biosensor for circulating tumor DNA based on carbon nanodots that emit methylene blue and red light.
The new sensor, according to the researchers, combines features of electrochemical and fluorescence sensors, whose signal sources and manufacturing processes are normally highly different.
An electrochemical sensor is a qualitative or quantitative method based on the correlation between concentration or other physical properties and the electrical signal change produced by the target.
A fluorescence sensor is a device that uses a specific target and recognition combination to transmit to a fluorescence element, changing the fluorescence's intensity or emission wavelength, to detect things qualitatively or quantitatively.
In addition to significantly increasing detection accuracy, integrating the two synchronous detection methods into a single, innovative system can also lessen the impact of background signals, instrument variations, and other variables on the signals that are collected. Peng Miao, a scientist from the Suzhou Institute of Biomedical Engineering and Technology and the study's principal investigator
In this sensor, Probe A is precisely immobilized at the electrode surface by tagging it at its 5' terminal. When the target is present, complete double strands are formed in Probe A between the target and its binding domain.
Probe B's second hairpin is also opened when the stem region is split apart and a single-stranded area is liberated. It had previously been connected to a red-emitting carbon nanodot's 3' terminal NH2. Probe C's hairpin structure is released once Probe B's single-stranded region is freed.
Additionally, the target sequence can be moved by Probe C to produce a complete three-way junction structure.
As a result, the target is employed again, resulting in several three-way intersections. Since there are several methylene blue molecules at the electrode contact at the 3' terminal of Probe C, it is possible to identify the target by recording a significant electrochemical response.
The released single-stranded swing arm formed between the 3' terminal of Probe A and the 5' terminal of Probe B on the three-way junction acts as the substrate, while the adjacent three-way junction's 3' terminal of Probe B serves as the DNAzyme. A coexisting structured DNA monolayer is produced as a result.
The release of the conjugated carbon nanodots into the solution and breakage of the substrate sequence are made possible by the presence of Mg2+.
According to Miao, "Original goal level may also be determined using fluorescence technology by detecting the increasing fluorescence emission."
Gel electrophoresis imaging and theoretical calculations were used to determine the process's viability. In a physiological environment, the synthesized carbon nanodots can maintain strong anti-interference and outstanding fluorescence stability.
Through a series of condition optimization and quantitative testing, Miao and his colleagues were able to develop the linear calibration curves of electrochemical and fluorescence intensities and target concentration. The linear range of these curves can be as large as six orders of magnitude.
It is also straightforward to determine the target's composition using fluorescence imaging. The dual-mode sensor developed in this work is novel, extremely sensitive, and scalable, making it an efficient tool for nucleic acid analysis and clinical diagnostics.





Comments
Post a Comment